Researchers have discovered that a rare type of genetic variant can be responsible for two well-known skeletal disorders.

Whereas many genetic conditions are caused by structural variations that involve deletions, insertions or duplications of segments of DNA, the scientists found that the genes were partially inverted – the DNA sequence was in the correct place, but flipped upside down. They have used the analogy of a book (see below).
The study, published in the BMJ’s Journal of Medical Genetics, was supported by the NIHR Oxford Biomedical Research Centre (BRC)
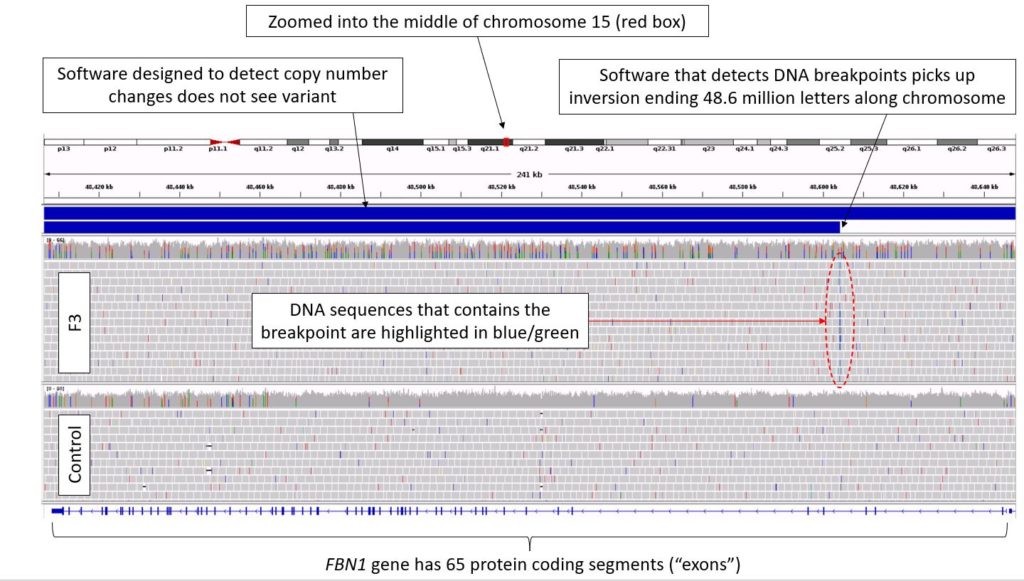
Most genetic testing methods are focused on identifying structural variants that alter the copy number; most genes come in pairs with one copy from each parent and so the copy number normally equals two. However, with inversions, the copy number remains the same, and so these variants have been easily missed.
The research team hope this discovery will lead to more people having the cause of their rare genetic conditions diagnosed.
Using data from the 100,000 Genomes Project, the researchers looked at 43 genes known to be connected to skeletal disorders. Ten people from three different families were found to have genetic inversions.
In two families, the gene that was inverted (GLI3) has been linked with Greig cephalopolysyndactyly syndrome. In another family, long suspected to have Marfan syndrome (a condition linked to tall stature, long spindly fingers and dilatation of the aorta), an inversion disrupting the FBN1 gene was identified.
Both FBN1 and GLI3 have long been linked to disease, but this particular type of genetic inversion has never been described before. The findings suggest that genetic testing for these conditions should be extended to include searching for these inversions.
The study’s lead author, Dr Alistair Pagnamenta, of the University of Oxford’s Wellcome Centre for Human Genetics, said: “These three families have had long diagnostic odysseys, and clinicians have suspected the gene responsible all along, given the characteristic symptoms. One of the families was tested several times over the course of many years by different labs and using multiple methods.
“The clinician who sees this family suspected it was Greig syndrome for almost 20 years, so she knew what they had clinically. But when the gene was tested it came back from the lab negative. The clinician sent it to multiple labs, and she was right – but it’s an unusual variant.”
Dr Pagnamenta added: “Because we have whole genome sequences, we can do a scan that can pick up these inversions. We’d like to see Genomics England and other labs to incorporate this kind of analysis, so that more families can have the diagnosis they deserve.”
Why genetic inversion is like a book
Imagine your genome is a book.
If a single letter is changed, you can search for it, like doing a spellcheck. Deletions are like ripping out several pages. This can be detected by weighing the book/genome.
But an inversion is like putting those ripped-out pages back in the book, but upside down. They can be difficult to detect: the book would weigh the same; and if you did a word count or a spellcheck, you wouldn’t be able to pick it up.
Genetic testing methods are very much biased towards structural variants that change copy number. The tests are not currently designed to look for inversions.
It would require us to flick through the ‘pages’ to see if one is upside down, next to one that is the ‘right-way-up’.
One question the Oxford BRC research team plan to look at in future: are inversions rare or just being missed?
They are now in the process of extending their analysis of 100,000 Genomes project samples to see if genome inversions are linked to other disease areas.